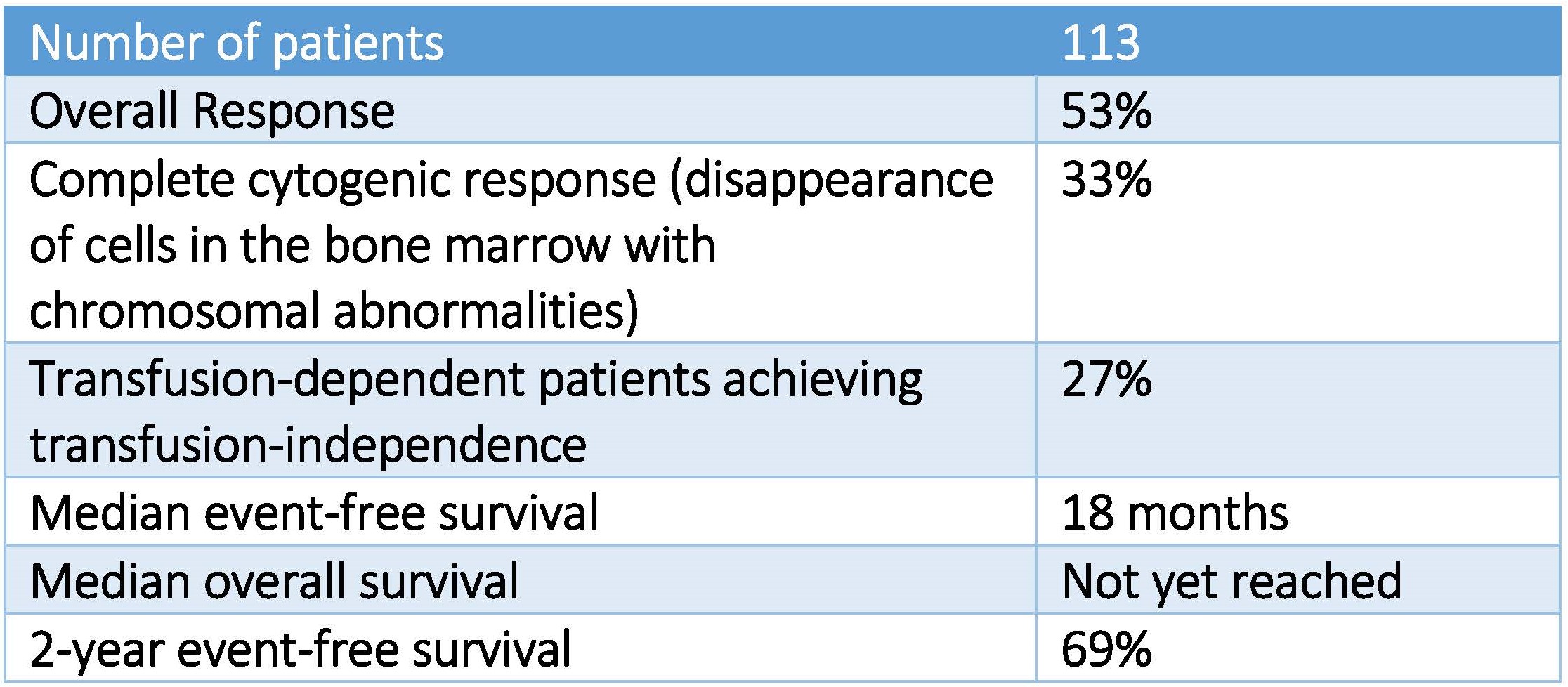
Results of studies in patients with low- and intermediate-risk myelodysplastic syndrome (MDS) treated with hypomethylating agents (low dose azacytidine or decitabine) followed for a median of 18 months were presented at the Society of Hematologic Oncology meeting in Houston on September 9, 2016.
What is Myelodysplastic Syndrome?
Myelodysplastic syndrome (MDS) is a clonal bone marrow disorder characterized by ineffective hematopoiesis such that the bone marrow doesn’t produce enough healthy blood cells, instead making too many immature cells or “blasts” (megakaryoblasts, proerythroblasts, myeloblasts, and lymphoblasts). These blasts die in the marrow or soon after entering the bloodstream, resulting in too few healthy blood cells and low blood counts. In its mildest form, MDS may be anemia, low platelets or low white blood count, but about 10% to 20% of diagnosed cases progress to acute myeloid leukemia (AML).
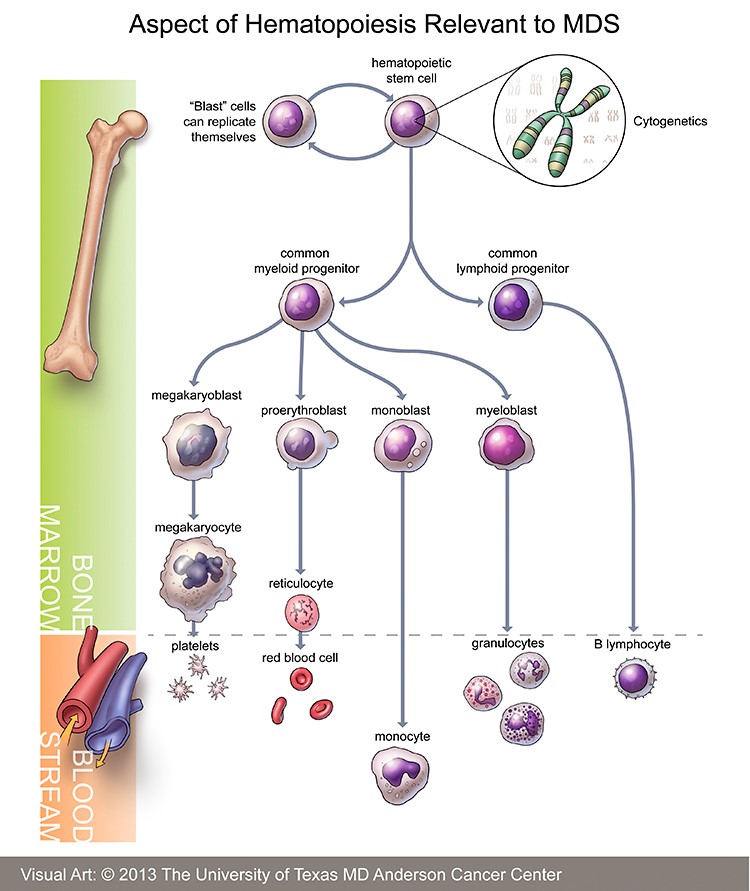
Hematopoiesis – https://www.mdanderson.org/content/dam/mdanderson/images/diseases/Anatomical%20Illustrations/Myelodysplastic_illustration.jpg
The myelodysplastic syndromes (MDS) encompass a series of hematologic conditions characterized by chronic cytopenias (anemia, neutropenia, thrombocytopenia) accompanied by abnormal cellular maturation. As a result, patients with MDS are at risk for symptomatic anemia, infection, and bleeding, as well as progression to acute myeloid leukemia (AML), which is often refractory to standard treatment.
MDS develops as the consequence of a series of genetic changes in a hematopoietic stem cell. These changes alter normal hematopoietic growth and differentiation, resulting in an accumulation of abnormal, immature myeloid cells in the bone marrow and the impairment of normal hematopoiesis. Advances in the identification of recurring chromosomal abnormalities and translocations have provided insight into the pathobiology of MDS.
The most common chromosomal abnormalities seen in MDS are del(5q), -7 or del(7q), trisomy 8, and del(20q). Deletion of the long arm of chromosome 5 (5q) is the most common chromosomal abnormality seen in MDS, occurring in approximately 15 percent of cases, overall. The RPS14 protein, required for the maturation of 40S ribosomal subunits, has been implicated in the genesis of the 5q- syndrome. Other genes located on 5q that are deleted in MDS, AML, or therapy-related MDS/AML with a del(5q) include EGR1, CTNNA1, and APC. Loss of function of Apc (adenomatosis polyposis coli gene, APC tumor suppressor gene) in animal models produces a condition simulating MDS and/or contributes to its development. Similarly, loss of function of EGR1 cooperates with mutations induced by alkylating agents to induce myeloid neoplasms in mouse models. These results provide support for the current model that the cooperative loss of multiple genes on 5q is required for the pathogenesis of myeloid disorders with a del(5q).
How does one develop MDS and what is the mortality?
Risk factors for myelodysplastic syndrome include:
- Age: MDS rarely occurs in people younger than 60
- Smoking tobacco
- Long-term exposure to chemicals, including benzene or other chemicals used in the petroleum and rubber industries
- Exposure to high levels of radiation, such a nuclear reactor accident or atomic bomb
- Prior chemotherapy or radiation therapy
- Inherited disorders, including: Fancomi anemia, Shwachman-Diamond syndromes, familial platelet disorder, and severe congenital neutropenia
The proportion of attributable deaths varied according to a patient’s MDS risk status. The proportion of deaths attributable to MDS increased from 31.5% among low-risk patients to 48.8% in the intermediate-risk category and 74.1% of patients with high-risk MDS. CAD/stroke as the cause of death decreased from a high of 26.3% of patients with low-risk disease to 20.6% of patients with intermediate risk-MDS to 9.3% of patients with high-risk MDS.
Treatment of Myelodysplastic Syndrome
Lower-risk myelodysplastic syndrome patients are treated initially for the specific complications of the disease, such as anemia and low blood counts. If more aggressive therapy is needed, strategies that are considered standard of care include chemotherapy using hypomethylating agents (5-azacitidine and decitabine) and lenalidomide.
Higher-risk myelodysplastic syndrome patients usually need more aggressive therapy, but much depends on the age and condition of the patient. Younger patients with high-risk disease are considered for front-line chemotherapy approaches followed perhaps by allogeneic stem cell transplantation.
Why do hypomethylation agents work in MDS?
Patients with MDS have hyper-methylated tumor suppressor genes p15 and p16, both of which inhibit cyclin D-CDK4/6. Tumor suppressor gene CDK1 is also hypermethylated in MDS – CDK1 encodes E-cadherin, which prevents epithelial mesenchymal transition. Epigenomic abnormalities silence the expression of these genes; hypomethylating agents reverse hypermethylation, thereby restoring tumor suppressor function.
Daskalakis demonstrated that reversal of p15 hypermethylation with decitabine was associated with clinical response:
We examined p15and p16 methylation status in bone marrow mononuclear cells from patients with high-risk MDS during treatment with decitabine, using a methylation-sensitive primer extension assay (Ms-SNuPE) to quantitate methylation, and denaturing gradient gel electrophoresis (DGGE) and bisulfite-DNA sequencing to distinguish individually methylated alleles. p15 expression was serially examined in bone marrow biopsies by immunohistochemistry. Hypermethylation in the 5′ p15 gene region was detected in 15 of 23 patients (65%), whereas the 5′ p16 region was unmethylated in all patients. Among 12 patients with hypermethylation sequentially analyzed after at least one course of decitabine treatment, a decrease in p15 methylation occurred in 9 and was associated with clinical response. DGGE and sequence analyses were indicative of hypomethylation induction at individual alleles. Immunohistochemical staining for p15 protein in bone marrow biopsies from 8 patients with p15 hypermethylation revealed low or absent expression in 4 patients, which was induced to normal levels during decitabine treatment. In conclusion, frequent, selectivep15 hypermethylation was reversed in responding MDS patients following treatment with a methylation inhibitor. The emergence of partially demethylated epigenotypes and re-establishment of normal p15 protein expression following the initial decitabine courses implicate pharmacologic demethylation as a possible mechanism resulting in hematologic response in MDS.
Aggerholm showed that hypermethylation predicts for poor response in patients with MDS:
The propensity of myelodysplastic syndrome (MDS) to transform into acute myeloid leukemia (AML) suggests the existence of common pathogenic components for these malignancies. Here, four genes implicated in the development of AML were examined for promoter CpG island hypermethylation in cells from 37 patients with different stages of MDS. Aberrant methylation was detected by polymerase chain reaction amplification of bisulfite-treated DNA followed by denaturing gradient gel electrophoresis. The highest rate of methylation was found for p15INK4B (51%), followed by HIC1 (32%), CDH1 (27%), and ER (19%). Concurrent hypermethylation of ≥3 genes was more frequent in advanced compared with early-stage MDS (P ≤ 0.05), and hypermethylation of p15INK4B was associated with leukemic transformation in early MDS (P ≤ 0.05). The median overall survival was 17 months for cases showing hypermethylation of ≥1 genes vs. 67 months for cases without hypermethylation (P = 0.002). Specifically, promoter hypermethylation identified a subgroup of early MDS with a particularly poor prognosis (median overall survival 20 months vs. 102 months; P = 0.004). In multivariate analysis including stage and thrombocyte count, hypermethylation of ≥1 genes was an independent negative prognostic factor (P < 0.05). These data suggest that hypermethylation of p15INK4B, HIC1, CDH1, and ER contribute to the development and outcome of MDS.
Decitabine is a natural analog of 2’-deoxy cytidine believed to exert its antineoplastic effects after phosphorylation and direct incorporation into DNA and inhibition of DNA methyltrans-ferase, causing hypomethylation of DNA and cellular differentiation or apoptosis. Decitabine inhibits DNA methylation in vitro, which is achieved at concentrations that do not cause major suppression of DNA synthesis. Decitabine-induced hypomethylation in neoplastic cells may restore normal function to genes that are critical for the control of cellular differentiation and proliferation. In rapidly dividing cells, the cytotoxicity of decitabine may also be attributed to the formation of covalent adducts between DNA methyltransferase and decitabine incorporated into DNA. Non-proliferating cells are relatively insensitive to decitabine.
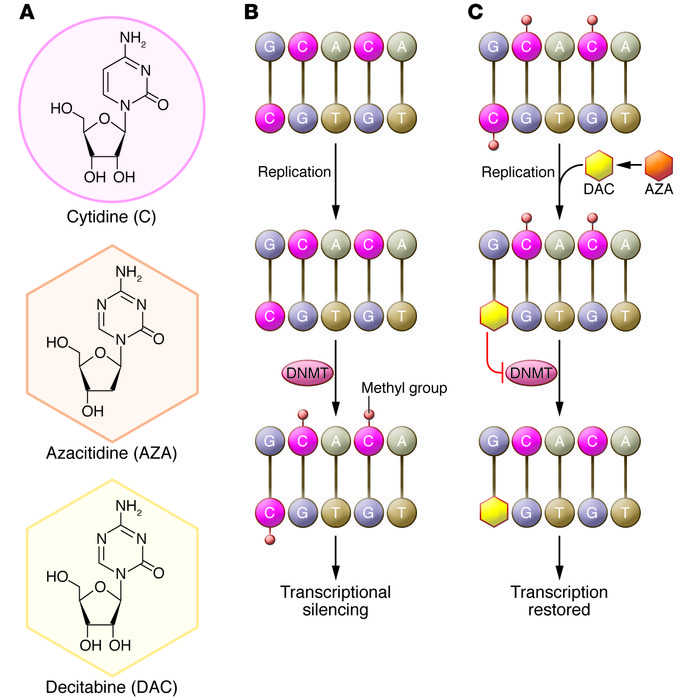
Biological mechanism of action of azacitidine (AZA) and decitabine (DAC).
(A) Structures of cytidine, azacitidine, and decitabine. (B) DNMTs methylate cytidine. Methylation of cytidine in gene promoter regions blocks the binding of transcription factors, leading to epigenetic silencing. DNMTs aberrantly methylate tumor suppressor genes in hematological malignancies and other cancers. (C) Azacitidine is converted to decitabine and is then incorporated into DNA in place of cytidine during replication. Both azacitidine and decitabine inactivate DNMTs, preventing gene methylation. The resultant hypomethylation leads to the transcription of previously silenced genes. Notably, azacitidine, but not decitabine, can be incorporated directly into RNA, inhibiting protein synthesis. https://www.jci.org/articles/view/69739/figure/1