The Sonic hedgehog (Hh) pathway is known to be important in basal cell carcinoma (BCC), which as an extremely common skin cancer that only rarely invades and metastasizes. An approved drug for patients with advanced BCC, Vismodegib (Erivedge), interferes with the Hedgehog pathway. The Patched (12 membrane-spanning receptor protein) normally disables Smoothened (7 membrane-spanning protein) rendering it functionally inert. This maintains transcription factor Gli in a cleaved state, acting as a transcriptional repressor. When Hedgehog proteins bind to Patched, Smoothened is released and protects Gli from cleavage. Uncleaved Gli travels to the nucleus and is an inducer of transcription, increasing Cyclin D1 and stimulating the cell cycle (proliferation). Vismodegib blocks the actions of Smoothened. It is administered orally in 150 mg capsules.
Researchers genetically profiled nearly 300 basal cell carcinoma (BCC) samples, and found additional recurrent mutations — including ones in MYCN, PTPN14, and LATS1 — that appear to drive disease. Basal cell carcinoma is one of the most common human cancers, affecting some 2 million people a year in the US, according to the American Academy of Dermatology, and is thought to be mainly caused by exposure to ultraviolet light.
Nikolaev, Antonarakis, and colleagues performed mutational screening on 293 BCC samples from 236 patients using a combination of matched tumor-normal whole-exome sequencing, targeted panel sequencing, and matched tumor-normal RNA-sequencing. The BCC samples, they noted, included 263 sporadic BCC cases and 30 Gorlin syndrome cases as well as vismodegib treatment-resistant, -sensitive, and -naïve cases. As expected, 85 percent of the BCCs the researchers examined had somatic mutations affecting the Hh pathway.
But by using an algorithm they developed that accounts for the background mutation rate per nucleotide, the researchers also uncovered 28 significantly recurrent mutations in 14 genes, including mutations in LATS1, SMO, MYCN, STK19, TP53, ERBB2, and PPP6C.
- Thirty percent of the BCCs harbored missense mutations in MYCN, an oncogene paralogous to MYC. Similar alterations, the researchers wrote, have been found in Burkitt lymphoma, where they were shown to interact with FBXW7, part of the complex that ubiquitinates Myc to target it for degradation.
- PTPN14, meanwhile, was mutated in nearly a quarter of the BCC samples studied. PTPN14 typically helps the translocation of YAP1, which is part of the Hippo signaling pathway, from the nucleus to the cytoplasm. That movement is hindered in cells with mutated PTPN14. This, the researchers noted, suggests that PTPN14 is a new tumor-suppressor gene in BCC that works by activating the Hippo-YAP pathway.
- The researchers also uncovered recurrent mutations in LATS1 and LATS2. LATS1 is a known tumor-suppressor kinase that’s activated by PTPN14 and phosphorylates YAP1 to prevent its translocation to the nucleus. The recurrent LATS1 mutation they uncovered is predicted to affect the protein’s structural stability and could affect its activity. Such LATS1-inactivating mutations, Antonarakis and his colleagues added, could be another way in which the Hippo-YAP1 pathway is activated in BCC.
A Closer look at PTPN14
The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP contains an N-terminal noncatalytic domain similar to that of band 4.1 superfamily cytoskeleton-associated proteins, which suggested the membrane or cytoskeleton localization of this protein. It appears to regulate lymphatic development in mammals, and a loss of function mutation has been found in a kindred with a lymphedema-choanal atresia.
Protein tyrosine phosphatase, which may play a role in the regulation of lymphangiogenesis, cell-cell adhesion, cell-matrix adhesion, cell migration, cell growth and also regulates TGF-beta gene expression, thereby modulating epithelial-mesenchymal transition. It mediates beta-catenin dephosphorylation at adhesion junctions and acts as a negative regulator of the oncogenic property of YAP, a downstream target of the hippo pathway, in a cell density-dependent manner.
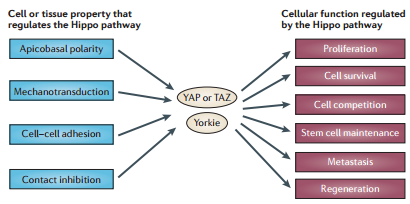
The Hippo Pathway and YAP
The Hippo pathway consists of a large network of proteins (at least 35 in both D. melanogaster and mammals have so far been identified) that control the growth of different tissues during development and regeneration, as well as in pathological states such as cancer. The heart of the Hippo pathway comprises a core kinase cassette that consists of a pair of related serine/threonine kinases, mammalian STE20-like protein kinase 1 (MST1; also known as STK4) and MST2 (also known as STK3), and large tumour suppressor 1 (LATS1) and LATS2, together with the adaptor proteins Salvador homologue 1 (SAV1) MOB kinase activator 1A (MOB1A) and MOB1B. These proteins limit tissue growth by facilitating LATS1- and LATS2-dependent phosphorylation of the homologous oncoproteins, Yes-associated protein (YAP; encoded by YAP1) and transcriptional co-activator with PDZ-binding motif (TAZ; also known as WWTR1).
Phosphorylation of YAP and TAZ represses their activity by creating 14‑3‑3 binding sites that cause them to accumulate in the cytoplasm and that also stimulate their ubiquitin-mediated proteolysis. YAP and TAZ, promote tissue growth and cell viability by regulating the activity of different transcription factors, including TEADs and SMADs.
- Uncontrolled cell proliferation is a fundamental aspect of neoplasia. Mutations in Hippo pathway proteins that lead to YKI hyperactivation in flies, or to YAP or TAZ hyperactivation in mammals, cause ectopic cell proliferation.
- In D. melanogaster imaginal discs, clones of cells with increased YKI activity progress through the cell cycle more rapidly than wild-type cells, and they proliferate precociously whereas wild-type cells quiesce.
- Besides quiescence, irreversible cell cycle exit (senescence) is an important tumour suppressor programme, and oncogene-induced senescence in cancer cells is dependent on the p53 and RB tumour suppressor pathways. A short hairpin RNA (shRNA) screen of kinases linked LATS2 to the RB tumour suppressor by showing that partial knockdown of LATS2 — thus partially relieving its suppression of YAP and TAZ — suppressed RB-induced senescence markers; furthermore, LATS2 overexpression enhanced RB-mediated silencing of E2F target genes.
- LATS2 has also been reported to activate p53 by binding to MDM2 and inhibiting its E3 ligase activity. In turn, p53 increases LATS2 transcription, which was shown to reinforce the G1/S checkpoint that is required for the arrest of cells that have become inappropriately tetraploid.
Collectively, these data suggest that unrestrained YAP and TAZ activity can counteract classical tumour suppressor checkpoints.
- Additionally, loss of the upstream Hippo pathway protein NF2, or overexpression of YAP, can provoke cells to overcome contact inhibition.
- Expression of the YAP and TAZ inhibitors LATS1 and LATS2 was significantly lower in metastatic, compared with non-metastatic, prostate cancers. In breast cancer, signal transduction from the metastasis suppressor leukaemia inhibitory factor receptor (LIFR) was shown to sequester and inactivate YAP; therefore, loss of LIFR expression could be one mechanism that results in YAP or TAZ hyperactivation during the metastasis of breast cancers. Loss of E-cadherin, which is intimately linked to EMT, might also cause YAP and TAZ de-repression in metastatic cells.
Therefore, de-regulation of the Hippo pathway has been shown to cause loss of contact inhibition, ectopic cell proliferation, accelerated cell cycle advancement, suppressed RB mediated silencing of E2F genes, blocking of senescence, and increased metastatic potential.
Additionally, the Hippo pathway activity is clearly perturbed through crosstalk with signaling pathways that frequently harbour oncogenic mutations. Multiple cancer-associated signaling networks engage in regulatory crosstalk with the Hippo pathway, often at the level of the YAP and TAZ oncoproteins. The WNT, transforming growth factor-β (TGFβ)–bone morphogenetic protein (BMP), Hedgehog (HH), Notch, insulin and mTOR pathways have all been reported to functionally interact with the Hippo pathway.
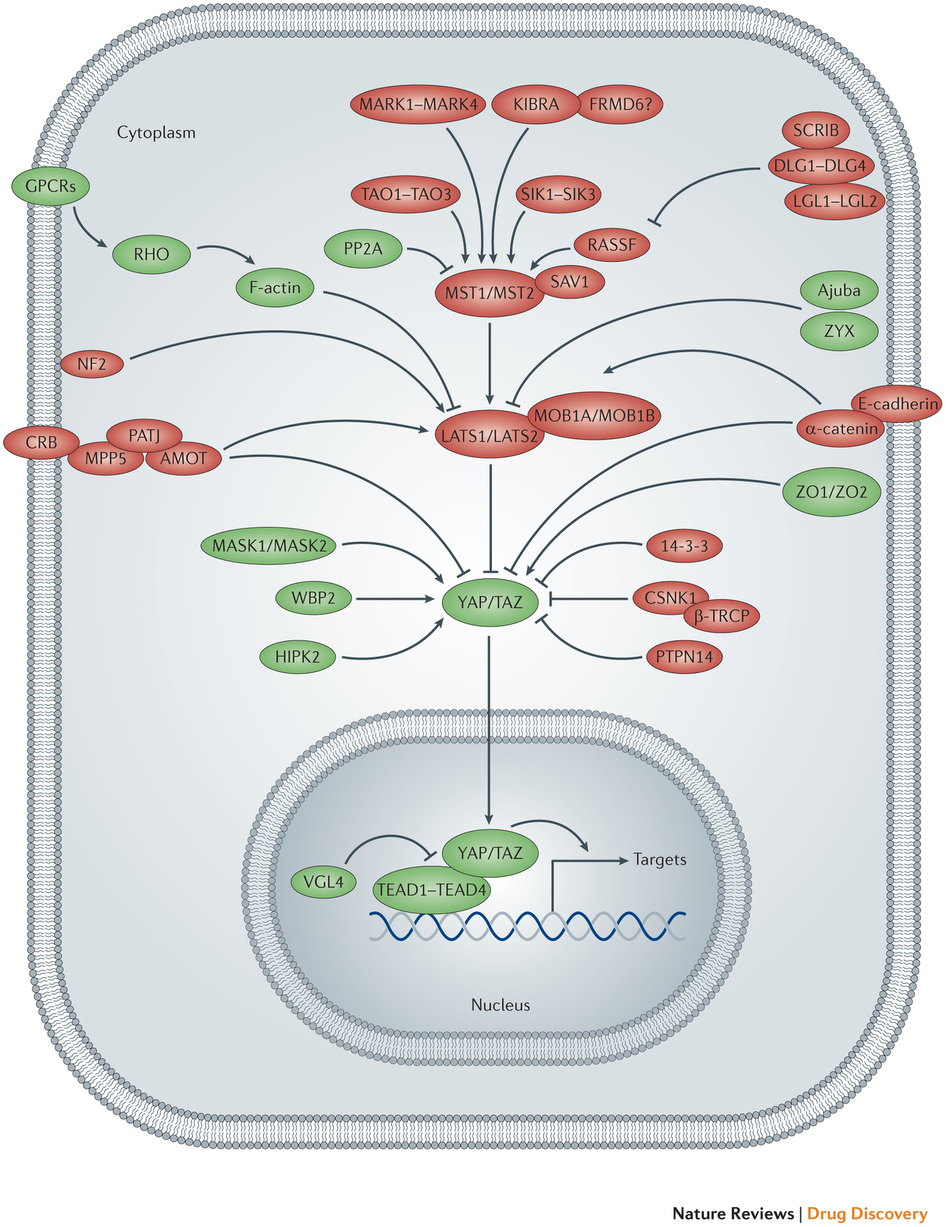
An outline of a cell is depicted, showing the nucleus and the Hippo pathway network. Mammalian Hippo pathway components that promote the activity of Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) are shown in green, whereas those that inhibit YAP and TAZ activity are shown in red. Pointed and blunt arrowheads indicate activating and inhibitory interactions, respectively. AMOT, angiomotin; β-TRCP, β-transducin repeat-containing E3 ubiquitin protein ligase; CSNK1, casein kinase 1; CRB, Crumbs homolog; DLG, discs large homolog; FRMD6, FERM domain-containing protein 6; GPCR, G protein-coupled receptor; HIPK, homeodomain-interacting protein kinase; KIBRA, kidney and brain protein; LATS, large tumour suppressor homolog; LGL, lethal giant larvae protein homolog; MARK, MAP/microtubule affinity-regulating kinase; MASK, multiple ankyrin repeats single KH domain-containing protein; MOB1A, MOB kinase activator 1A; MPP5, membrane protein, palmitoylated 5 (also known as PALS1); MST, mammalian STE20-like protein kinase; NF2, neurofibromin 2 (also known as Merlin); PATJ, PALS1-associated tight junction protein; PP2A, protein phosphatase 2A; PTPN14, protein tyrosine phosphatase, non-receptor type 14; RASSF, RAS association domain-containing family protein; SAV1, Salvador homolog 1; SCRIB, Scribble homolog; SIK, salt-inducible kinase; TAO, thousand and one amino acid protein kinase; TEAD, TEA domain-containing sequence-specific transcription factor; VGL4, vestigial-like protein 4; WBP2, WW domain-binding protein 2; ZO, zona occludens protein; ZYX, Zyxin protein.
If phosphorylation sequesters and neutralizes YAP/TAZ, how does PTPN14, which is a phosphatase, repress oncogenic activity of YAP?
Given that PTPN14 is a phosphatase, and that LATS1 and LATS2 neutralize YAP and TAZ via phosphorylation, which leads to the non-nuclear localization, How does PTPN14 modulate YAP?
Exactly how YAP1 localization is regulated under various conditions remains to be further defined. It was proposed that when cells reach confluence, cell–cell interaction and cytoskeleton rearrangement trigger a cascade of cell signaling to activate the hippo pathway. Two components in this pathway, MST1/2 and LATS1/2, get activated and coordinate to phosphorylate YAP1 at the Ser127 site. The 14-3-3 protein recognizes the phosphorylated YAP1 and sequesters it in the cytoplasm, which is the suggested mechanism for regulating YAP1 by cell density via the hippo pathway (Zhao et al. 2007). In recent years, several groups, including us, have reported that angiomotin family proteins AMOT, AMOTL1, and AMOTL2 can retain YAP1 in cytosol through a direct protein–protein interaction that occurs independently of YAP1 phosphorylation (Chan et al. 2011; Wang et al. 2011; Zhao et al. 2011; Oka et al. 2012). Since the association between PTPN14 and YAP1 is similar to that between angiomotin protein family members and YAP1, we asked whether PTPN14 could also translocate YAP1 from the nucleus to the cytoplasm.
Exogenously expressed PTPN14 mostly localized in the cytoplasm and partially colocalized with actin filaments at the plasma membrane (Figure panel A). Endogenous YAP1 showed mostly nuclear staining with mild cytoplasm localization in the sparse cells (i.e., low density) (Figure panel A). However, when PTPN14 was overexpressed, we observed a dramatic translocation of YAP1 from the nucleus to the cytoplasm. This translocation was independent of PTPN14 phosphatase activity, as the phosphatase catalytic-dead PTPN14 mutant (PTPN14C1121S) (Barr et al. 2006) could still translocate YAP1 to the cytoplasm. However, the two PY motif deletion mutant of PTPN14 (PTPN14delPY1/2), which disrupts the association of PTPN14 with YAP1, failed to translocate YAP1 to the cytoplasm. Collectively, these findings suggest that PTPN14 can mediate the translocation of YAP1 from the nucleus to the cytoplasm via their physical interaction and thereby inhibit YAP1 transcriptional functions.
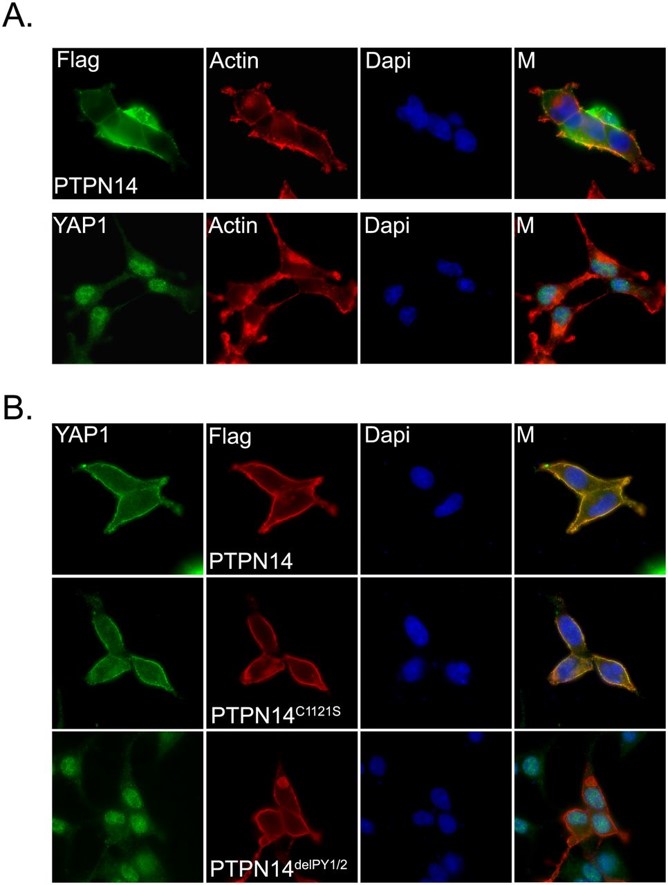
PTPN14 induces translocation of YAP1 from the nucleus to the cytoplasm. (A) Cellular localization of PTPN14 and YAP1 in HEK293 cells. Cells were transfected with constructs encoding SFB-tagged PTPN14. The cellular localization of PTPN14 was detected by anti-Flag immunostaining. The localization of endogenous YAP1 was detected by immunostaining using anti-YAP1 antibody. Actin filaments were labeled with TRITC-conjugated phalloidin. Nuclei were stained by DAPI. (M) Merged. (B) PY motifs of PTPN14 were required for YAP1 translocation. The localization of endogenous YAP1 was detected by anti-YAP1 antibody in HEK293 cells expressing the indicated SFB-tagged PTPN14 mutants. The localizations of SFB-tagged PTPN14 mutants were detected with anti-Flag antibody. Nuclei were stained by DAPI. (M) Merged. http://genesdev.cshlp.org/content/26/17/1959.full
To address whether the binding to YAP1 is required for PTPN14 function in acini formation, we reintroduced shRNA-resistant PTPN14, its phosphatase-dead mutant (PTPN14C1121S), or its two PY motif-deleted mutant (PTPN14delPY1/2) in PTPN14-depleted MCF10A cells using a doxycycline-inducible system. Expression of either wild-type PTPN14 or its phosphatase-dead mutant reduced aberrant acini formation caused by the loss of endogenous PTPN14 and restored the normal acini structures. However, re-expression of the two PY motif-deleted mutant of PTPN14 could not rescue the aberrant acini formation in these cells. These data confirmed that the direct inhibition of YAP1 by PTPN14 is required for normal acini formation of MCF10A cells, which depends on a physical protein–protein interaction between PTPN14 and YAP1 but not on PTPN14 phosphatase activity.
Hippo Pathway and tissue architecture
Epithelial and other tissue structures manifest a highly ordered spatial architecture. Over the past decade, our knowledge has greatly increased of how upstream regulatory proteins feed into the Hippo pathway to control its activity. Strikingly, most of these upstream proteins also control fundamental properties of tissues such as ABCP (apicobasal cell polarity), planar cell polarity (PCP) and the adhesion of cells to other cells, as well as to the extracellular matrix. Thus, Hippo pathway activity is tightly coupled to tissue and cellular architecture and function. Derangement of tissue architecture is a fundamental feature of solid human tumours, and thus might underlie the deregulated Hippo pathway activity that has been reported in different human cancers.