Celgene and Nurix entered into a collaboration to develop small molecule treatments for cancer that target the ubiquitin proteasome system (UPS).
What is Ubiquitin?
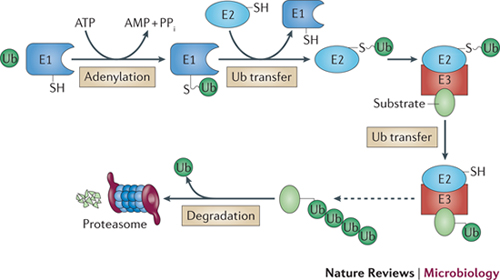
Ubiquitin is a small protein composed of 76 amino acids, and is highly conserved and ubiquitously expressed in the organisms of eukaryotic kingdom. The ubiquitin proteasome pathway first processes the attachment of ubiquitin to a target protein which involves three critical enzymes, the ubiquitin activating enzyme or E1 enzyme, the ubiquitin conjugating enzyme or E2 enzyme, and the ubiquitin ligase or E3 ligase. In most cases, ubiquitin tagged target proteins are subjected to degradation, in which the protein complexes are recognized by the 26S proteasome, a large multienzyme complex to mediate protein degradation. This whole process called ubiquitination which includes following three critical steps: firstly, ubiquitin needs to be activated from its precursor by adding to E1 through an ATP-dependent manner, and activated ubiquitin is then transferred to the ubiquitin-conjugating enzyme E2; secondly, E2 interacts with E3 to identify the substrate, and by which ubiquitin is attached to the target protein; and thirdly, the ubiquitin conjugated protein is recognized by the 26S proteasome, and through which, the target protein is degraded to small peptides or amino acids by the proteasome enzymes. Of note, ubiquitin can be released by the deubiquitinating enzymes (DUBs), and therefore, ubiquitin conjugation to target substrates is a reversible process.
How does the UPS work?
Ubiquitylation is a common signal for eukaryotic 26S proteasomes and involves a cascade of E1 ubiquitin-activating, E2 ubiquitin-conjugating and E3 ubiquitin ligase enzymes. In this cascade, E1 (plus ATP) first adenylates the carboxy-terminal carboxylate of ubiquitin (Ub), forming Ub–AMP, and then forms a Ub thioester intermediate (E1–Ub). Ubiquitin is transferred from E1 to E2, and then to the protein target with assistance from E3 (although ubiquitylation without E3 can occur118). Typically, an isopeptide bond is formed between the ubiquitin C-terminal carboxylate and the ɛ-amino group of a Lys side chain of the substrate protein or the growing ubiquitin chain (Lys48-linked ubiquitin chains are common signals for 26S proteasomes). Deubiquitylating enzymes within 26S proteasomes release and recycle ubiquitin during substrate protein degradation. PPi, inorganic pyrophosphate.
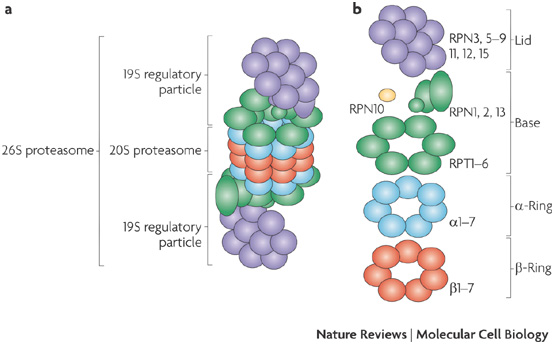
a | The 26S proteasome consists of the catalytic 20S proteasome (a barrel of four stacked rings: two outer -rings and two inner -rings) and the 19S regulatory particle (RP, also known as PA700). b | Subunit composition of the 26S proteasome. The regulatory particle is further divided into the base and the lid subcomplexes, which are composed of regulatory particle triple-A (RPT) and regulatory particle non-ATPase (RPN) subunits. RPN10 is colored yellow because it is supposed to be located at the base–lid interface. http://www.nature.com/nrm/journal/v10/n2/fig_tab/nrm2630_F1.html
Why is the UPS Important?
UPS is critical in the appropriate function of (1) cell cycle control, (2) DNA damage response via p53, and (3) the apoptosis cascade.
- Control of cell cycle involves three key classes of regulatory molecules, cyclin, cyclin-dependent kinases (CDKs) and cyclin-dependent kinase inhibitors (CKIs). Ubiquitin-mediated proteolysis of these regulatory molecules is one of the key mechanisms underlying cell cycle control.
- Given the role of p53 played in preventing genome mutation, it has been considered as “the guardian of the genome.” Although p53 is subject to a variety of post-translational modifications, ubiquitination of p53 has emerged as a fundamental regulatory mechanism. Studies revealed that p53 can be modified by a number of E3 ubiquitin ligases such as Pirh2, COP1, ARF binding protein and E6AP, while the murine double minute 2 (MDM2) oncoprotein, however, is the most critical negative regulator for p53 activity and the most extensively studied p53 E3 ligase. Under physiological condition, the cells only maintain low levels of p53, which is controlled by the rapid degradation of p53 via poly-ubiquitination, primarily mediated by the high basal levels of MDM2. MDM2 acts as the major E3 ubiquitin-protein ligase to interact with p53, and by which it represses p53 transcriptional activity by mediating its ubiquitination and proteasomal degradation. In contrast, p53 undergoes a significant increase in protein stability upon exposing to the DNA damage inducing factors such as stressful insults. It is believed that DNA damage stabilizes p53 in part via the DNA damage signaling pathway that implicates the sensor kinases such as the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related protein (ATR) kinase, and the effector kinases. The signals generated by these kinases lead to the dissociation of the p53/MDM2 complex along with the activation of p53. Once activated, p53 induces the transcriptional regulation of a variety of genes to arrest cell cycle, a process necessary for DNA damage repair. Nevertheless, when DNA damage is beyond the extent of cellular repair capacity, p53 would then induce apoptosis to prevent the malignant transformation of cells.
- For TNF-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis signaling, there are multiple mechanisms by which apoptosis are regulated by the ubiquitin–proteasome pathway. First, proteasome inhibitors can decrease Fas-like inhibitor protein (FLIP) protein levels in tumors, resulting in increased apoptosis signaling due to increased caspase-8 activation. This appears to involve the ubiquitin ligase TNF receptor activation factor-2 (TRAF2) and acts indirectly by causing cell-cycle arrest at a stage where there is high degradation of the FLIP–TRAF2 complex. Second, the regulation of the proapoptotic Bcl-2 family member BAX occurs indirectly. Apoptosis signaling and caspase activation results in a confirmation change in the normally monomeric BAX, which exposes the BH3 domain of BAX, leading to dimerization and resistance to ubiquitin degradation. BAX then translocates into the mitochondria, resulting in the release of pro-apoptotic mitochondrial factors such as cytochrome cand second mitochondria-derived activator of caspase (SMAC). This results in the activation of caspase-9 and formation of the apoptosome and efficient apoptosis signaling. A third mechanism of the regulation of TRAIL signaling in the ubiquitin–proteasome pathway is mediated by the inhibitor of apoptosis proteins (IAP) E3 ligases. These IAPs can directly bind to caspases but also can act as ubiquitin ligases for caspases, resulting in the degradation of these caspases. IAP binding to caspases can be inhibited by SMAC, which exhibits a caspase-9 homology domain. The fourth mechanism for apoptosis activation by proteasome inhibitors is through the stabilization of the inhibitor of the B (IB)/NF-B complex and prevention of nuclear translocation of the anti-apoptosis transcription factor NF- During TRAIL-DR4, DR5 signaling, this pathway is activated by interactions of activated Fas-associated death domain with activated receptor-interacting protein (RIP), which in turn activates NF-B-inducing kinase and phosphorylates IB. Therefore, the inhibition of IB degradation blocks this RIP-mediated anti-apoptosis signaling event.
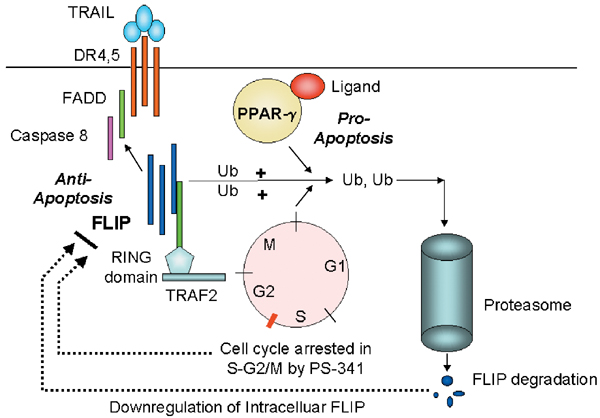
Ubiquitination of FLIP regulates TRAIL-induced apoptosis. The trimeric TRAIL results in the recruitment of death domain receptor 4 or death domain receptor 5 (DR4, DR5) molecules. The TRAIL-induced conformational changes of the DR4/DR5 death domain result in the recruitment of FADD and procaspase-8, which then signals apoptosis. FLIP contains a death domain that can compete with caspase-8 cytoplasmic binding domain of DR4, DR5. The intracellular levels of FLIP are regulated by the proteasome degradation pathway. One proposed pathway is binding of FLIP to TRAF2. TRAF2 contains a RING zinc-finger domain and possesses E3 ligase activity.
What is Nurix doing?
Nurix is targeting E2 conjugating enzymes and E3 ligases that control the UPS. Regulation of protein stability by the UPS is modulated by the activity of these E2 and E3 enzymes which control the tagging of proteins with ubiquitin for degradation by the proteasome. The human genome encodes approximately 1,000 different E3 ligases and 60 E2 enzymes. To date, a number of these ligases have recently been shown to play key roles in human diseases, particularly in cancer.
Approved Drugs Targeting the UPS
Discovered prior to scientific understanding of the central role of the UPS in cellular metabolism, highly successful drugs such as Velcade®, Kyprolis® and Revlimid® have been found to act through modulation of the UPS. Velcade® and Kyprolis® are 26S proteasome inhibitors and Revlimid® is a modulator of cereblon, an E3 ligase. And yet, these products target only two of the multitude of UPS pathways now available for drug discovery.
The UPS in Cancer
Some tumors have altered regulation of the UPS preventing degradation of proteins encoded by oncogenes that drive tumor cell growth. This dysregulation can be caused by specific mutations at the interface of an E3 ligase and its oncogenic substrate. Nurix identifies compounds that selectively re-engage the UPS to target these harmful proteins.
Conversely, some tumors hyper-activate particular ubiquitin ligases through gene amplification or over-expression, triggering inappropriate degradation of protective tumor suppressor proteins. In such cases, selective inhibitors of these ligases may restore tumor suppressor activity.
Selective modulators of certain E3 ligases that have been identified as checkpoints for the immune system.