Tazemetostat is a small molecule inhibitor of EZH2 being developed by Epizyme, Inc for the treatment of patients with Non-Hodgkin Lymphoma, including germinal center and non-germinal center diffuse large B-cell lymphoma and follicular lymphoma, as well as genetically-defined solid tumors. In many human cancers, misregulated EZH2 enzyme activity results in misregulation of genes that control cell proliferation—without these control mechanisms, cancer cells are free to grow rapidly.
Tazemetostat may also be a potential treatment for patients with INI1-deficient or SMARCA4-negative tumors resulting from the misregulation of EZH2, including a cancer known as malignant rhabdoid tumor (MRT), a rare and deadly form of cancer that primarily affects very young children, and synovial sarcoma which occurs in children and young adults.
Wild-type EZH2 catalytic activity in the formation of germinal centers in non-diseased lymph nodes is critical for B-lymphocyte maturation. The importance of EZH2 to B-cell lymphoma likely lies in its ability to regulate B-cell differentiation. Excellent responses in 9 of 15 patients in a Phase 1 study were reported.
EZH2 (Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit) is a histone methyltransferase that trimethylates histone 3 lysine 27 (H3K27me3) for gene repression. EZH2 gene encodes a member of the Polycomb-group (PcG) family. PcG family members form multimeric protein complexes, which are involved in maintaining the transcriptional repressive state of genes over successive cell generations. This protein associates with the embryonic ectoderm development protein, the VAV1 oncoprotein, and the X-linked nuclear protein. This protein may play a role in the hematopoietic and central nervous systems. Multiple alternatively spliced transcript variants encoding distinct isoforms have been identified for this gene.
EZH2 inhibition initiates a differentiation program that enables lymphoma cells to proceed through the normal processes of B-cell selection, growth regulation and maturation, which is critical for the treatment of B-cell lymphomas. In epithelial cancers, EZH2 is central to the EMT – epithelial-mesenchymal transition.
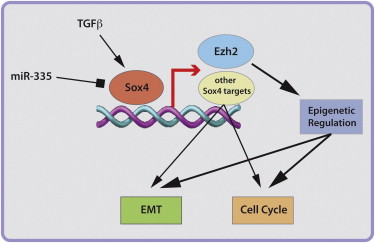
Gene expression profiling has uncovered the transcription factor Sox4 with upregulated activity during TGF-β-induced epithelial-mesenchymal transition (EMT) in normal and cancerous breast epithelial cells. Sox4 is indispensable for EMT and cell survival in vitro and for primary tumor growth and metastasis in vivo. Among several EMT-relevant genes, Sox4 directly regulates the expression of Ezh2, encoding the Polycomb group histone methyltransferase that trimethylates histone 3 lysine 27 (H3K27me3) for gene repression. http://www.genecards.org/cgi-bin/carddisp.pl?gene=EZH2
Ablation of Ezh2 expression prevents EMT, whereas forced expression of Ezh2 restores EMT in Sox4-deficient cells. Ezh2-mediated H3K27me3 marks associate with key EMT genes, representing an epigenetic EMT signature that predicts patient survival. Our results identify Sox4 as a master regulator of EMT by governing the expression of the epigenetic modifier Ezh2. EZH2 can also methylate non-histone proteins such as the transcription factor GATA4 and the nuclear receptor RORA.
INI1 and SMARCA4 are subunits of a regulatory complex, SWI/SNF, that opposes the enzymatic function of EZH2. Due to a variety of genetic alterations, INI1 can lose its regulatory function. As a result, EZH2 activity is misregulated, causing EZH2 to play a driving, oncogenic role in a set of genetically defined cancers that include malignant rhabdoid tumors and synovial sarcoma. Hence, tazemetostat, an EZH2 inhibitor is being developed for the treatment of tumors deficient in SWI/SNF functioning.
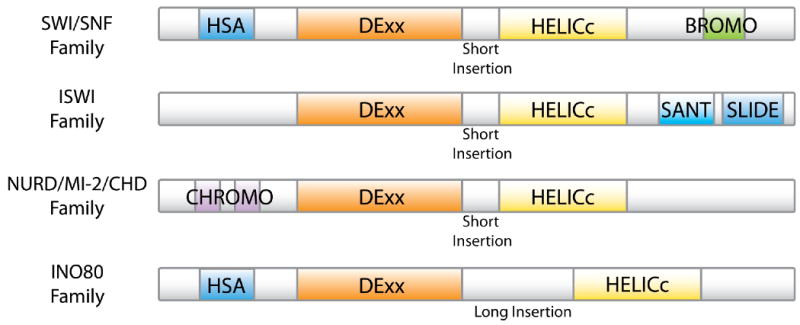
Classification of ATP-dependent chromatin remodeling complexes: The ATPase subunit of all the remodeling complexes belongs to the superfamily II helicase group. The ATPase always contains a DExx and a HELICc domain, spaced by a linker. The remodelers are classified into different families based on the presence of additional domains on their ATPase subunits. The SWI/SNF family contains a HSA (helicase/SANT-associated) domain, involved in actin binding, and a bromodomain important for the binding of acetylated lysines. The ISWI family contains the SANT and SLIDE domains, important for histone binding. The CHD/NURD/Mi-2 family contains a tandem Chromo domain, also used for histone binding. The INO80 family, like the SWI/SNF family, comprises a HSA domain but it is also characterized by the presence of a longer insertion between the DExx and the HELICc domains. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2924208/
Epigenetic regulation is responsible for gene expression by controlling the conditions under which genes are transcribed. Epigenetic regulation is carried-out by HAT (histone acetyltransferases), HDAC (histone deacetylase) and HMT (histone methyltransferases). Since these enzymes regulate patterns of gene expression by targeting selected genes or chromosome regions, the therapeutic potential of such inhibitors is enormous, particularly for treating cancers or other diseases showing aberrant patterns of gene expression.

