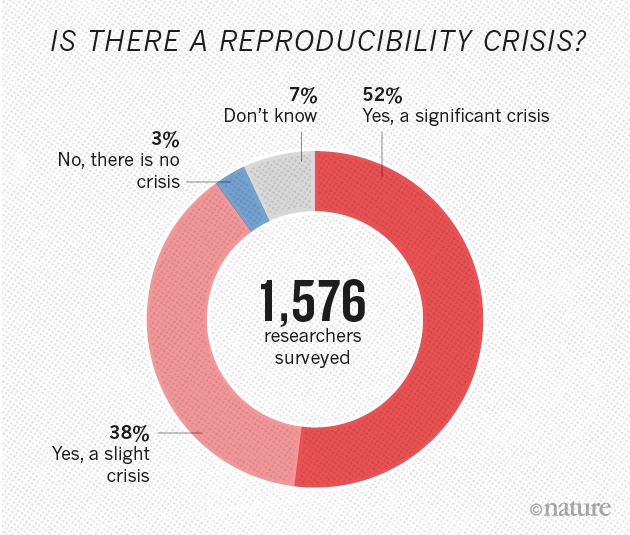
When is a brain really dead? The answer to this question was made far more complicated by the recent work of Dr. Nenad Sestan and colleagues at Yale University. Their astonishing paper entitled “Restoration of brain circulation and cellular functions post-mortem,” appeared in the journal Nature[1] this April. In it, the authors were able to demonstrate that brains taken from slaughtered animals (pigs in this case), could be “reanimated” 4 hours later in the laboratory – and made at least partially functional for some 6 hours thereafter. I use the word reanimated with some trepidation – it implies the brains were dead and somehow brought back to life. That is not quite the story – rather, the organ turns out to be far more resilient than we previously realized and the research team simply identified a way to tap into that inherent resiliency.
A brief description of the research study: 32 brains from slaughtered pigs were delivered to the research team on ice. Within 4 hours, the scientists carefully perfused the brains using a proprietary surgical procedure, pumping apparatus, and oxygen and nutrient-rich solution collectively termed BrainEx. The team then analyzed the brains for specific cellular, metabolic, and electrical activities. What they found was incredible. The BrainEx perfusion system restored a number of brain functions, including glucose and O2 utilization and concomitant CO2 production (indicative of metabolic function), induced inflammatory responses (suggesting an active immune system), active microcirculation (evidence of structural integrity), and electrical activity (with neuronal firing).
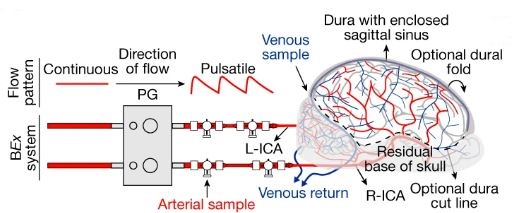
With respect to the last point, it should be noted the investigators were well aware of the ethical concern that full restoration of brain function could potentially lead to a state of “consciousness.” What it would mean for a disembodied brain to attempt to operate without peripheral sensory input, and what the organ might remember, was simply too much to consider; the brains were treated pharmacologically to assure no coordinated higher level cognitive activity was possible. Said another way, the renewed brains could not begin to think.
Overall, the results suggested to a first approximation, functional activity of the otherwise dead brain had been restored. Importantly from an experimental standpoint, control perfusates were without effect – the brains degenerated much like untreated specimens.
Against the backdrop of these remarkable results are the inevitable ethical questions that follow. Returning to the one posed above – when is a brain really dead? If our understanding of brain death requires a deeper examination, how then do we determine legally, ethically, and/or spiritually, when a person truly dies? What are the implications of this work on the issue of organ donations? Is a brain death as currently defined sufficient to permit the harvesting of a person’s organs? Will (and should) donations ebb as people begin to question the legitimacy of declarations of death. Also, could future brain reanimation experiments include restoration of conscious thought? Certainly the ability to control the brain in this manner permits the testing of drugs in new ways – potentially impacting several health scourges of our time including Alzheimer’s disease, Parkinson’s disease, and other age-related neurodegenerative disorders.
Naturally, science requires replication, expansion, and more careful delineation of what is, and is not possible with the technology described. But what enormous doors have been opened for neuroscientists, neurologists, neuropharmacologists, cognitive scientists, and the many others interested in the structure and function of the brain. I suspect ethicists will also be very busy sorting through these and the inevitable follow-up studies – especially as applications involving the human brain are contemplated.
SRT – July 2019
[1] Vrselja Z, et al., Nature 568, 336-343 (2019). PMID: 30996318